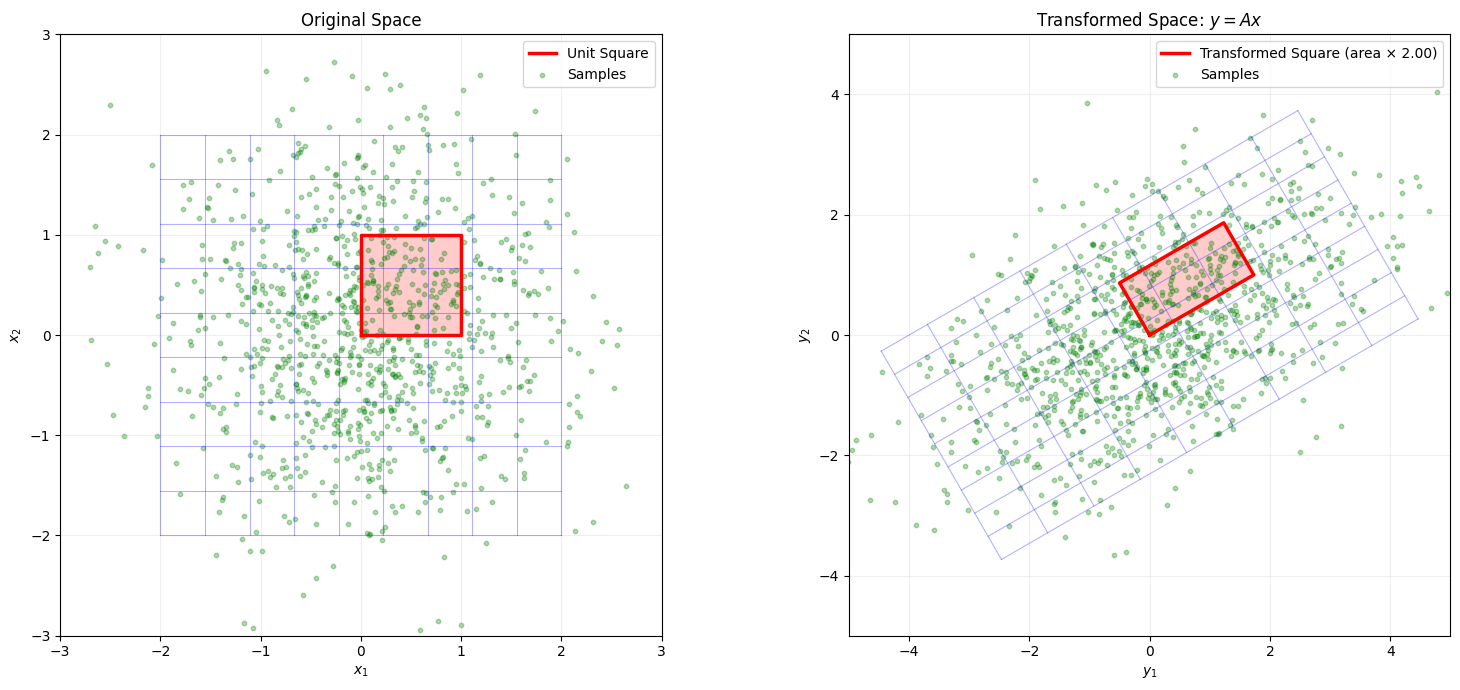
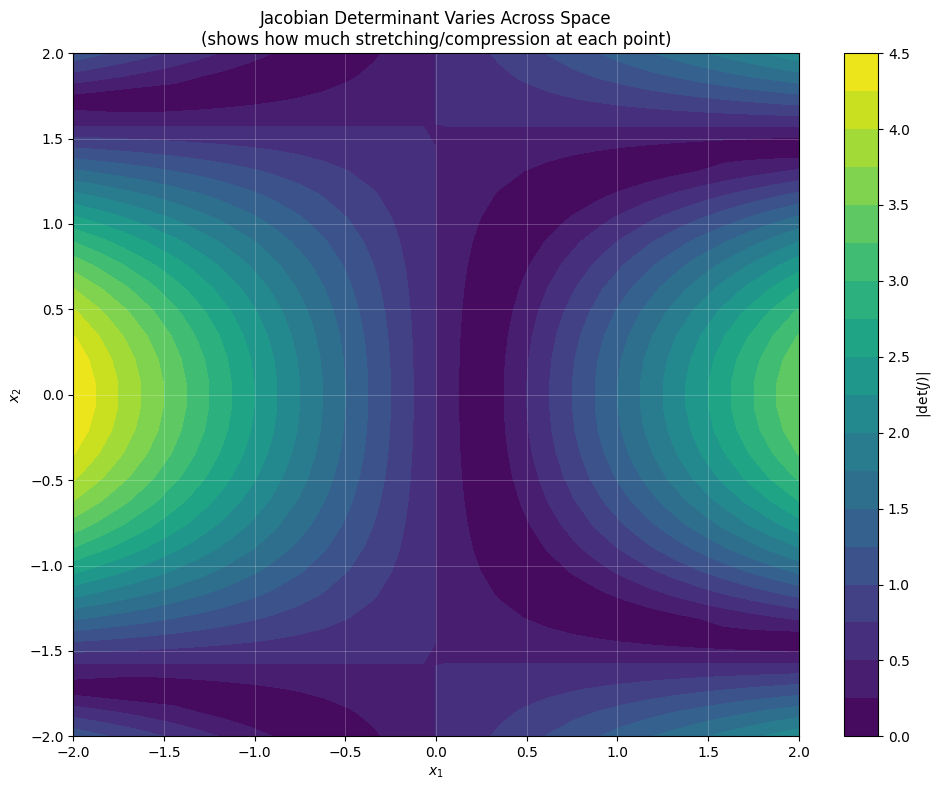
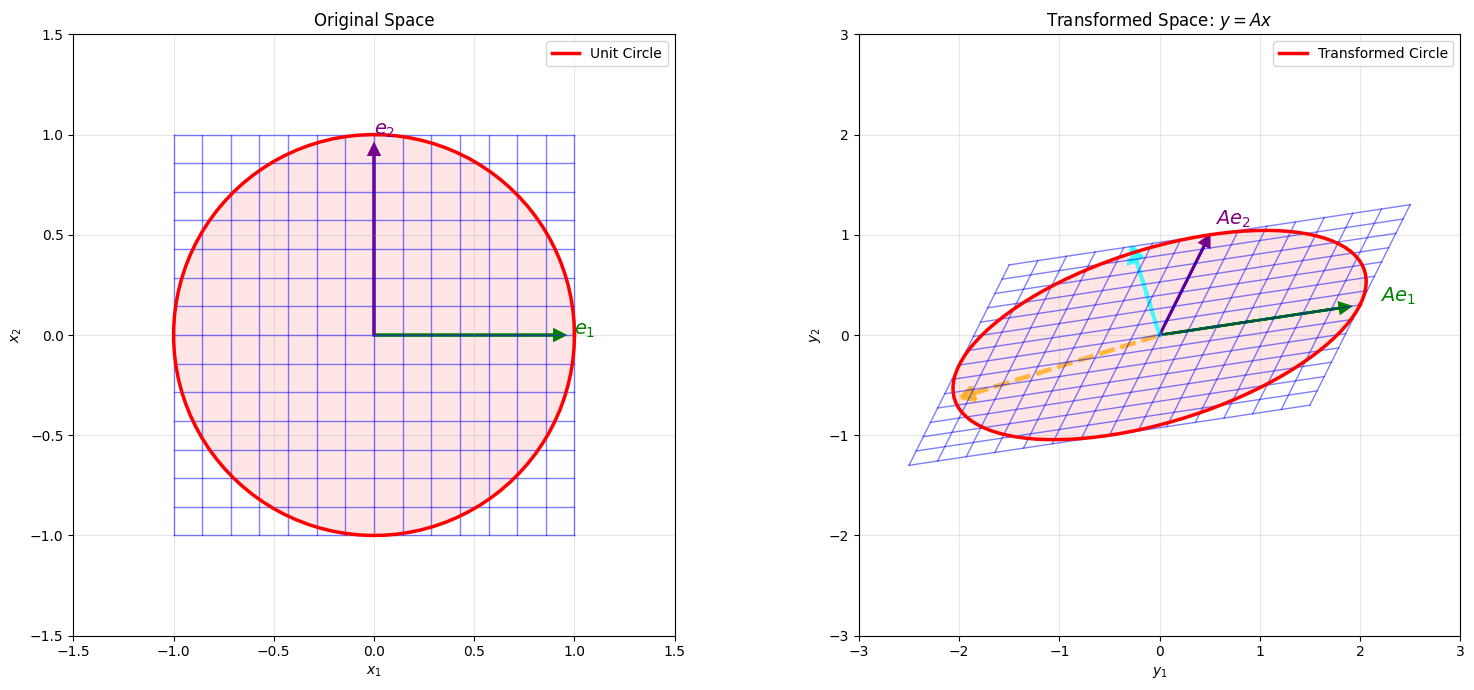
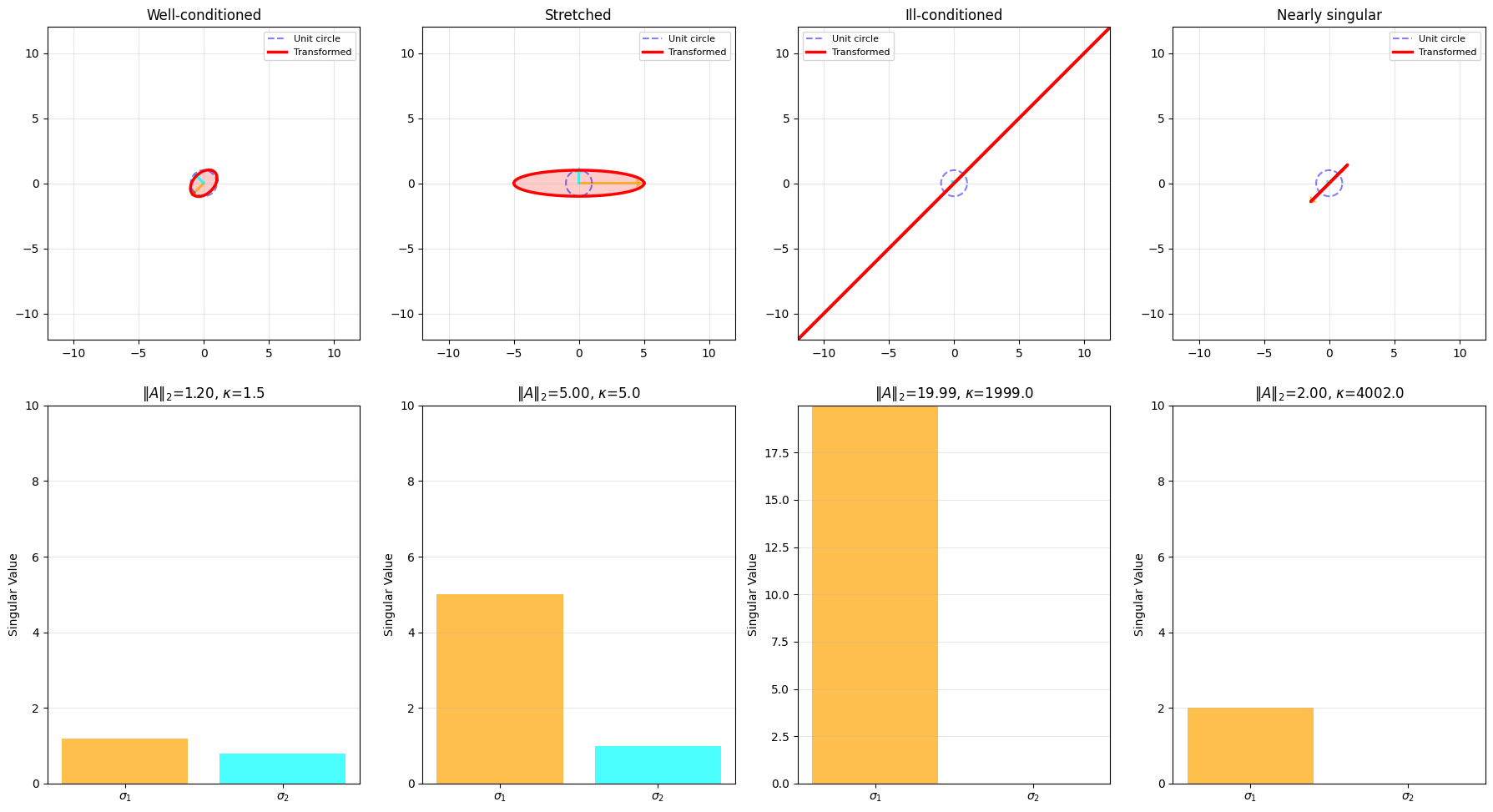
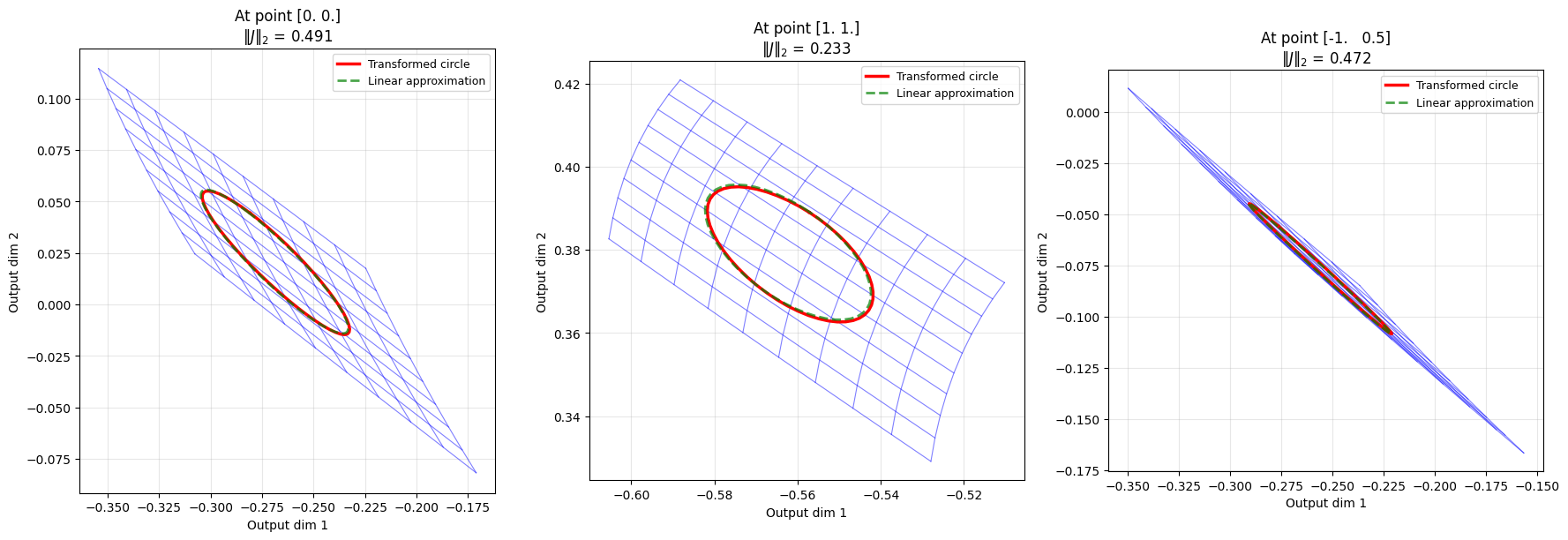
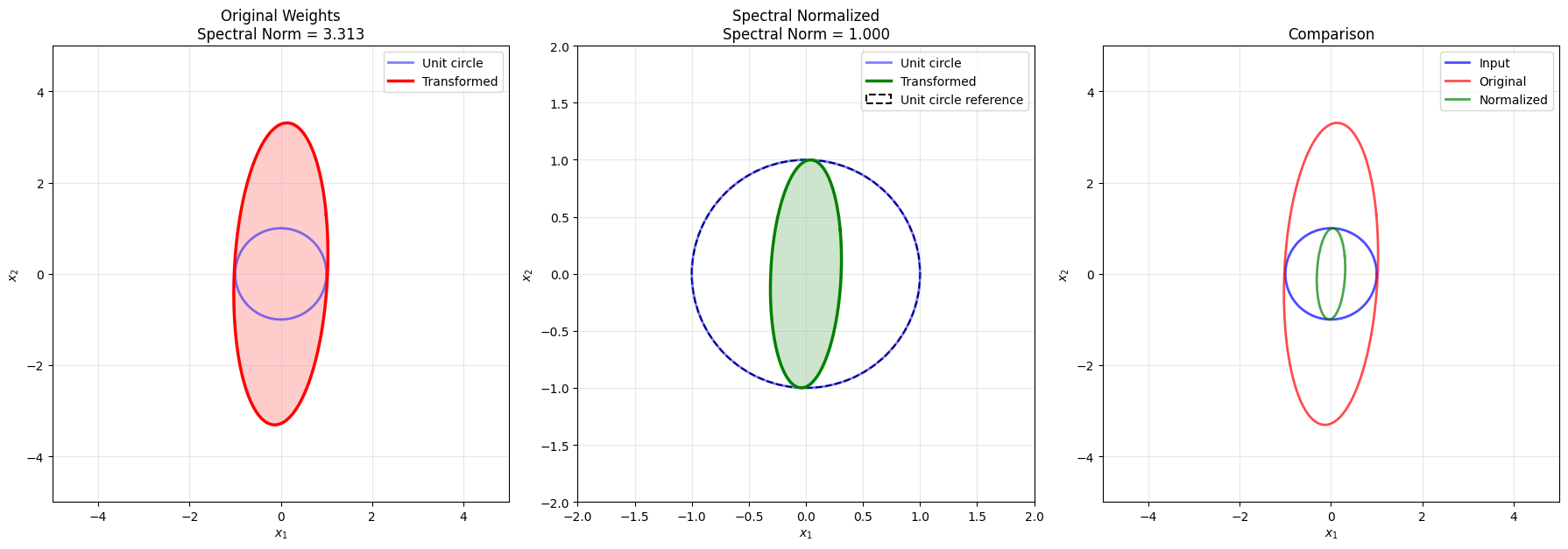
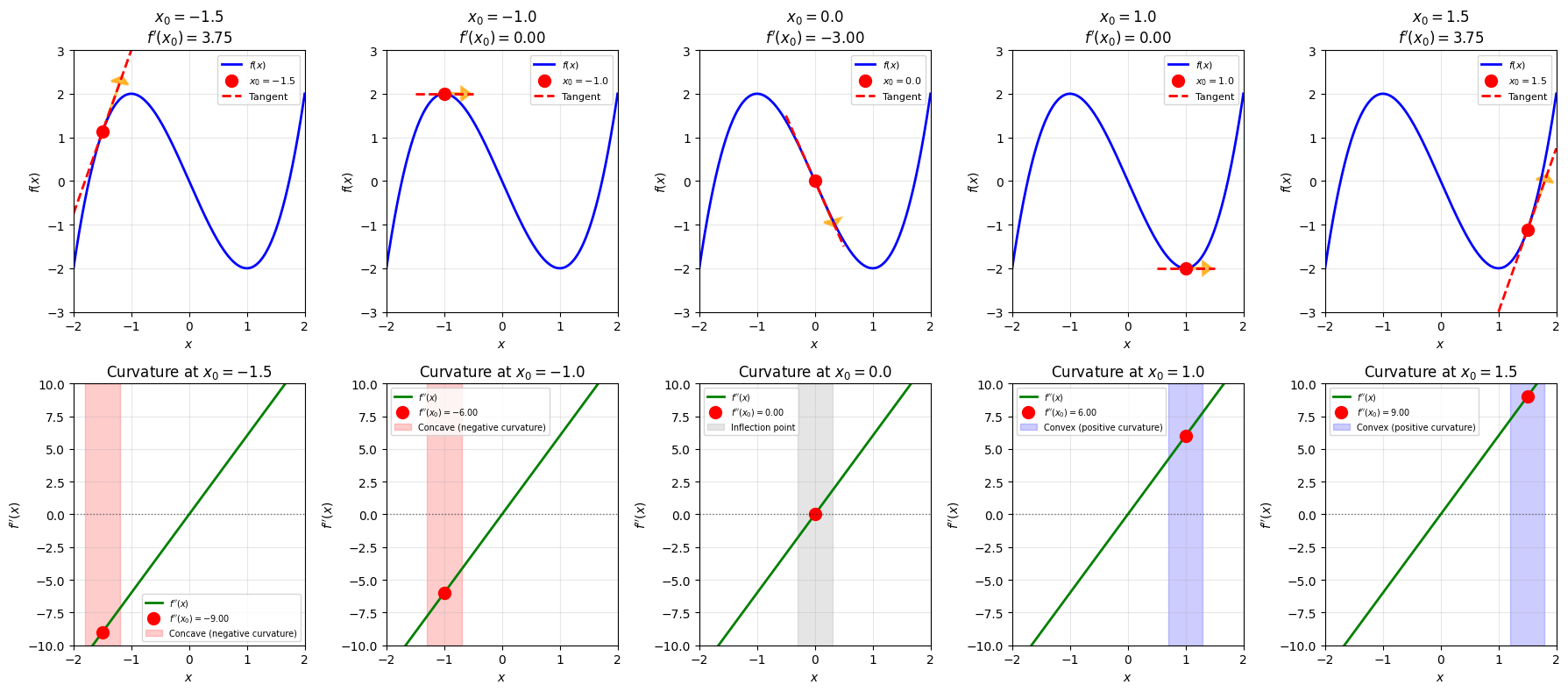
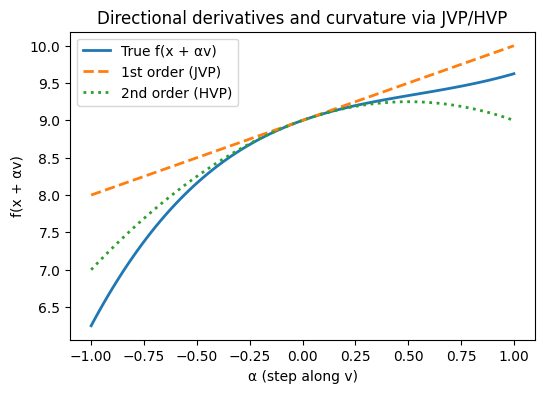
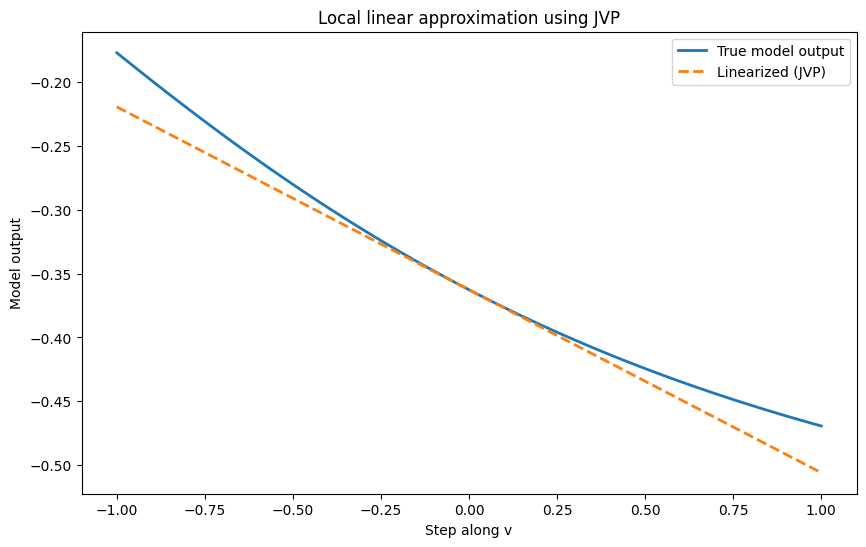
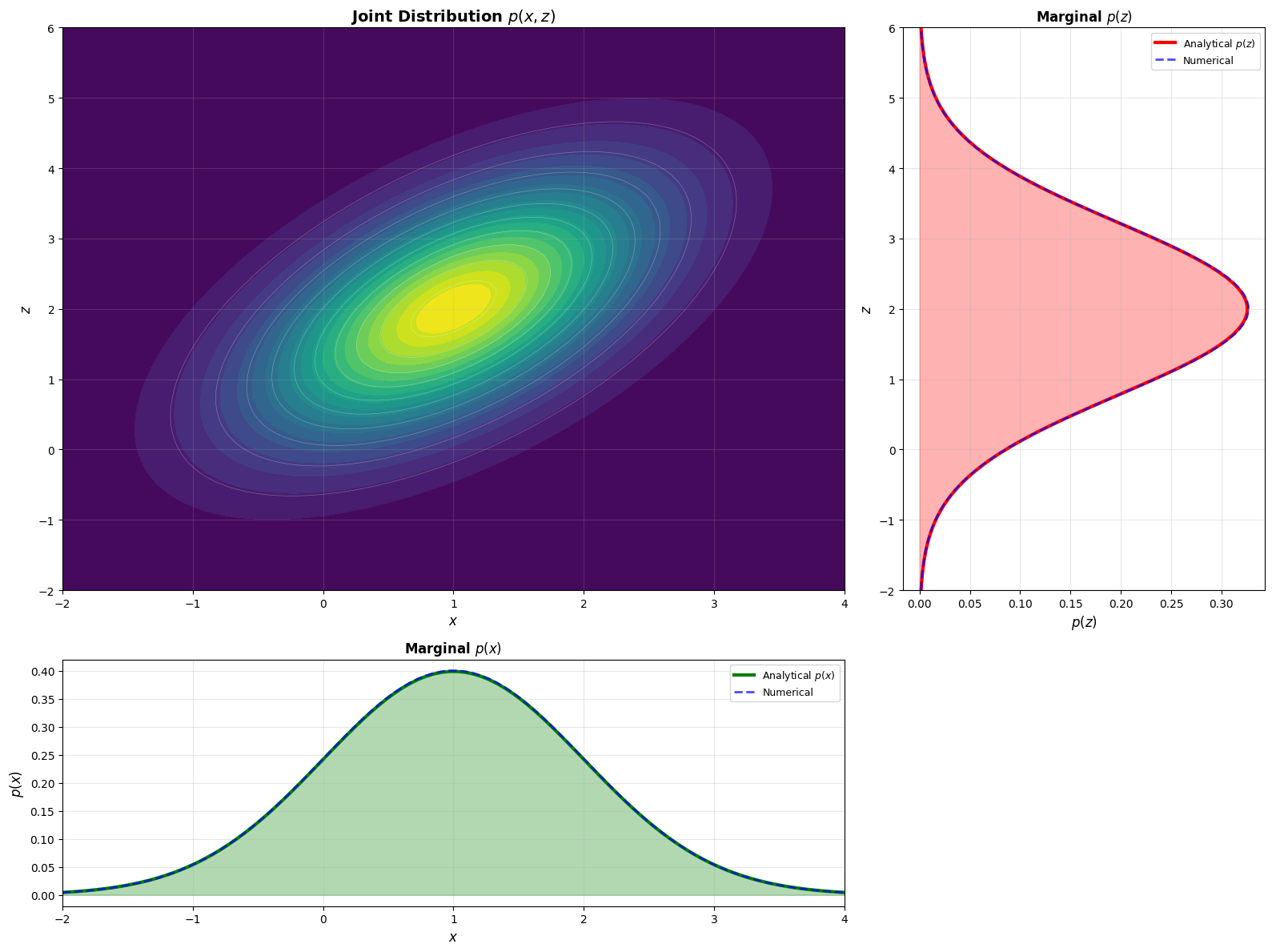
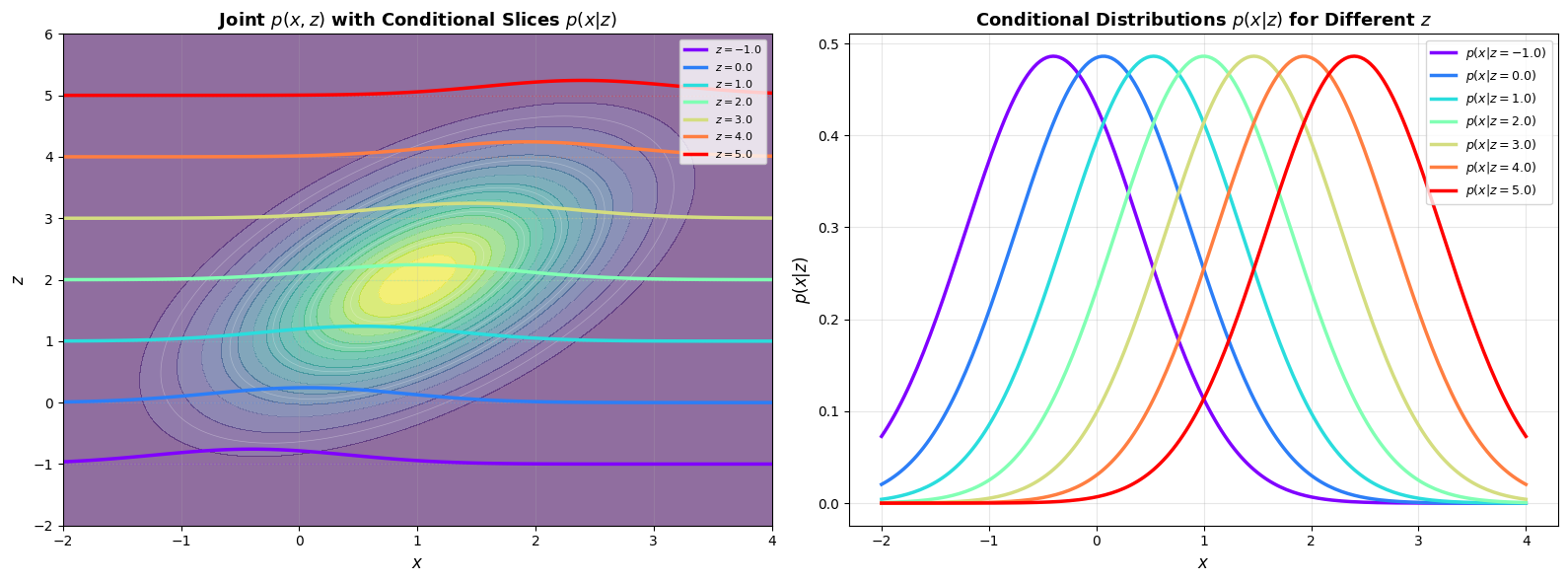
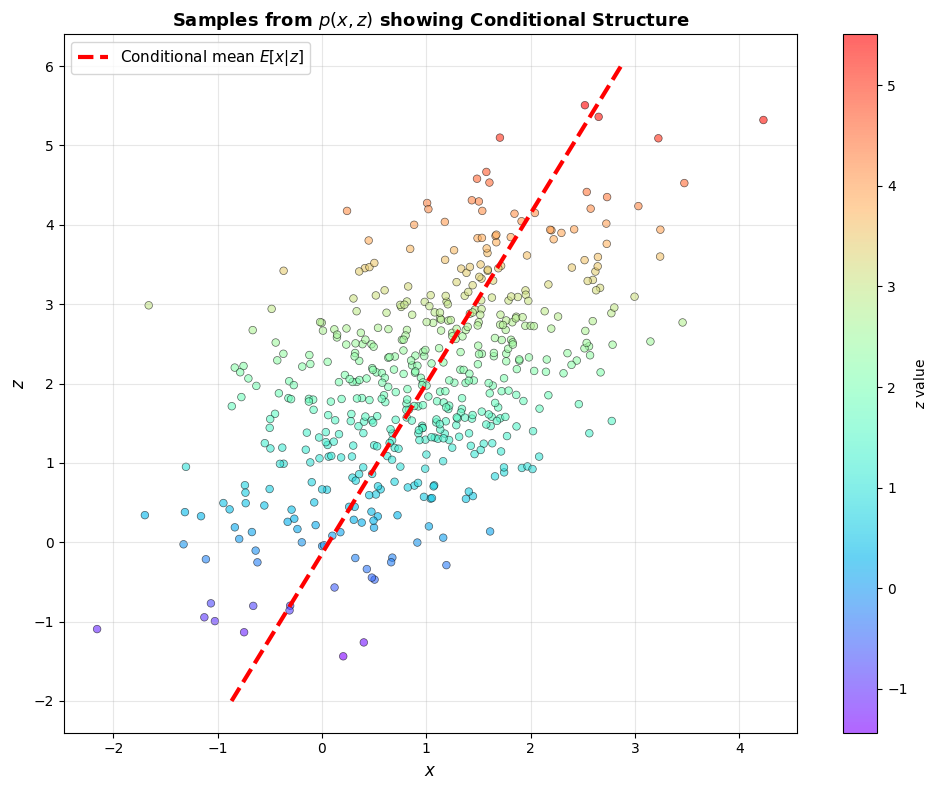
# Create an interesting transformation matrix
# Mix of rotation and stretching
np.random.seed(42)
A = np.array([[2.0, 0.5],
[0.3, 1.0]])
print("Transformation matrix A:")
print(A)
# Compute SVD
U, singular_values, Vt = np.linalg.svd(A)
print(f"\nSingular values: {singular_values}")
print(f"Spectral norm (max singular value): {singular_values[0]:.4f}")
print(f"Condition number: {singular_values[0]/singular_values[1]:.4f}")
# Create a grid of points
n_grid = 15
x = np.linspace(-1, 1, n_grid)
y = np.linspace(-1, 1, n_grid)
X, Y = np.meshgrid(x, y)
# Create points on grid
points = np.stack([X.ravel(), Y.ravel()], axis=0)
# Apply transformation
transformed_points = A @ points
# Reshape back to grid
X_trans = transformed_points[0, :].reshape(X.shape)
Y_trans = transformed_points[1, :].reshape(Y.shape)
# Create visualization
fig = plt.figure(figsize=(16, 7))
# Left: Original grid
ax1 = fig.add_subplot(121, aspect='equal')
for i in range(n_grid):
ax1.plot(X[i, :], Y[i, :], 'b-', alpha=0.5, linewidth=1)
ax1.plot(X[:, i], Y[:, i], 'b-', alpha=0.5, linewidth=1)
# Draw unit circle
theta = np.linspace(0, 2*np.pi, 100)
unit_circle_x = np.cos(theta)
unit_circle_y = np.sin(theta)
ax1.plot(unit_circle_x, unit_circle_y, 'r-', linewidth=2.5, label='Unit Circle')
ax1.fill(unit_circle_x, unit_circle_y, 'red', alpha=0.1)
# Draw coordinate axes
ax1.arrow(0, 0, 0.9, 0, head_width=0.05, head_length=0.05, fc='green', ec='green', linewidth=2)
ax1.arrow(0, 0, 0, 0.9, head_width=0.05, head_length=0.05, fc='purple', ec='purple', linewidth=2)
ax1.text(1.0, 0, '$e_1$', fontsize=14, color='green', fontweight='bold')
ax1.text(0, 1.0, '$e_2$', fontsize=14, color='purple', fontweight='bold')
ax1.set_xlim(-1.5, 1.5)
ax1.set_ylim(-1.5, 1.5)
ax1.set_xlabel('$x_1$')
ax1.set_ylabel('$x_2$')
ax1.set_title('Original Space')
ax1.legend()
ax1.grid(True, alpha=0.3)
# Right: Transformed grid
ax2 = fig.add_subplot(122, aspect='equal')
for i in range(n_grid):
ax2.plot(X_trans[i, :], Y_trans[i, :], 'b-', alpha=0.5, linewidth=1)
ax2.plot(X_trans[:, i], Y_trans[:, i], 'b-', alpha=0.5, linewidth=1)
# Transform unit circle -> ellipse
ellipse_points = A @ np.stack([unit_circle_x, unit_circle_y])
ax2.plot(ellipse_points[0, :], ellipse_points[1, :], 'r-', linewidth=2.5,
label='Transformed Circle')
ax2.fill(ellipse_points[0, :], ellipse_points[1, :], 'red', alpha=0.1)
# Draw transformed coordinate axes
e1_trans = A @ np.array([1, 0])
e2_trans = A @ np.array([0, 1])
ax2.arrow(0, 0, e1_trans[0]*0.9, e1_trans[1]*0.9,
head_width=0.1, head_length=0.1, fc='green', ec='green', linewidth=2)
ax2.arrow(0, 0, e2_trans[0]*0.9, e2_trans[1]*0.9,
head_width=0.1, head_length=0.1, fc='purple', ec='purple', linewidth=2)
ax2.text(e1_trans[0]*1.1, e1_trans[1]*1.1, '$Ae_1$',
fontsize=14, color='green', fontweight='bold')
ax2.text(e2_trans[0]*1.1, e2_trans[1]*1.1, '$Ae_2$',
fontsize=14, color='purple', fontweight='bold')
# Draw singular vectors
# Right singular vectors (V) point in direction of maximum stretch
# Left singular vectors (U) point in direction of output
v1 = Vt[0, :] # Direction of max stretch in input
v2 = Vt[1, :] # Direction of min stretch in input
u1 = U[:, 0] # Direction in output
u2 = U[:, 1]
# Draw principal directions in transformed space
sigma1_vec = singular_values[0] * u1
sigma2_vec = singular_values[1] * u2
ax2.arrow(0, 0, sigma1_vec[0]*0.9, sigma1_vec[1]*0.9,
head_width=0.15, head_length=0.15, fc='orange', ec='orange',
linewidth=3, alpha=0.7, linestyle='--')
ax2.arrow(0, 0, sigma2_vec[0]*0.9, sigma2_vec[1]*0.9,
head_width=0.15, head_length=0.15, fc='cyan', ec='cyan',
linewidth=3, alpha=0.7, linestyle='--')
ax2.set_xlim(-3, 3)
ax2.set_ylim(-3, 3)
ax2.set_xlabel('$y_1$')
ax2.set_ylabel('$y_2$')
ax2.set_title('Transformed Space: $y = Ax$')
ax2.legend()
ax2.grid(True, alpha=0.3)
plt.tight_layout()
plt.show()