import numpy as np
from matplotlib import pyplot as plt
import seaborn as sns
from sklearn.datasets import fetch_openml
from datetime import datetime, timedelta
import pandas as pd
from pandas.plotting import register_matplotlib_converters
register_matplotlib_converters()
sns.set_context('notebook')
# Fetch the data
mauna_lao = fetch_openml('mauna-loa-atmospheric-co2', as_frame = False)
print(mauna_lao.DESCR)
data = mauna_lao.data
# Assemble the day/time from the data columns so we can plot it
d1958 = datetime(year=1958,month=1,day=1)
time = [datetime(int(d[0]),int(d[1]),int(d[2])) for d in data]
X = np.array([1958+(t-d1958)/timedelta(days=365.2425) for t in time]).T
X = X.reshape(-1,1) # Make it a column to make scikit happy
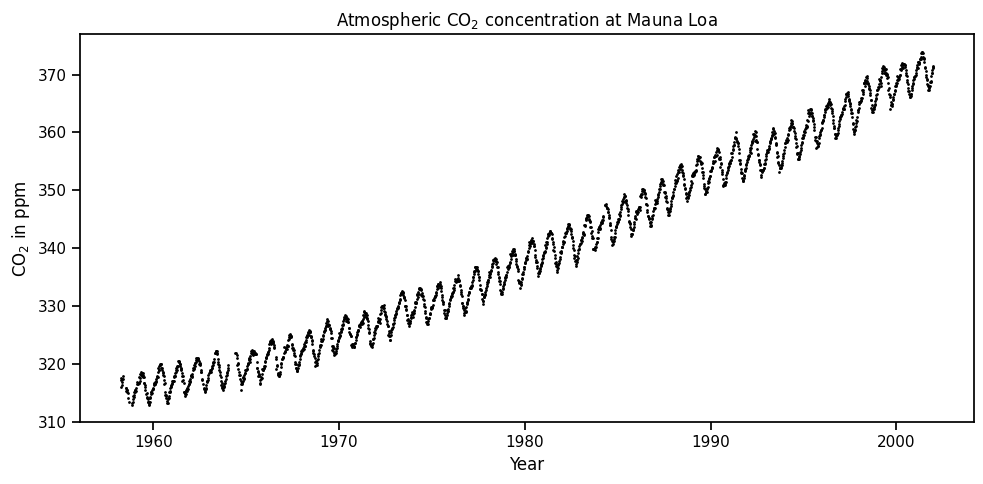
y = np.array(mauna_lao.target)**Weekly carbon-dioxide concentration averages derived from continuous air samples for the Mauna Loa Observatory, Hawaii, U.S.A.**<br><br>
These weekly averages are ultimately based on measurements of 4 air samples per hour taken atop intake lines on several towers during steady periods of CO2 concentration of not less than 6 hours per day; if no such periods are available on a given day, then no data are used for that day. The _Weight_ column gives the number of days used in each weekly average. _Flag_ codes are explained in the NDP writeup, available electronically from the [home page](http://cdiac.ess-dive.lbl.gov/ftp/trends/co2/sio-keel-flask/maunaloa_c.dat) of this data set. CO2 concentrations are in terms of the 1999 calibration scale (Keeling et al., 2002) available electronically from the references in the NDP writeup which can be accessed from the home page of this data set.
<br><br>
### Feature Descriptions
_co2_: average co2 concentration in ppvm <br>
_year_: year of concentration measurement <br>
_month_: month of concentration measurement <br>
_day_: day of month of concentration measurement <br>
_weight_: number of days used in each weekly average <br>
_flag_: flag code <br>
_station_: station code <br>
<br>
**Author**: Carbon Dioxide Research Group, Scripps Institution of Oceanography, University of California-San Diego, La Jolla, California, USA 92023-0444 <br>
**Source**: [original](http://cdiac.ess-dive.lbl.gov/ftp/trends/co2/sio-keel-flask/maunaloa_c.dat) - September 2004
Downloaded from openml.org.