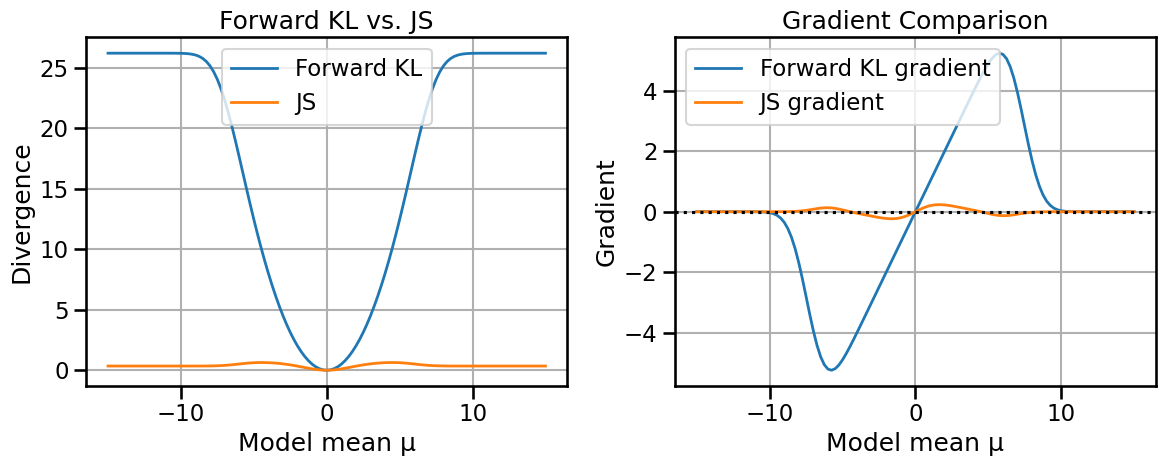
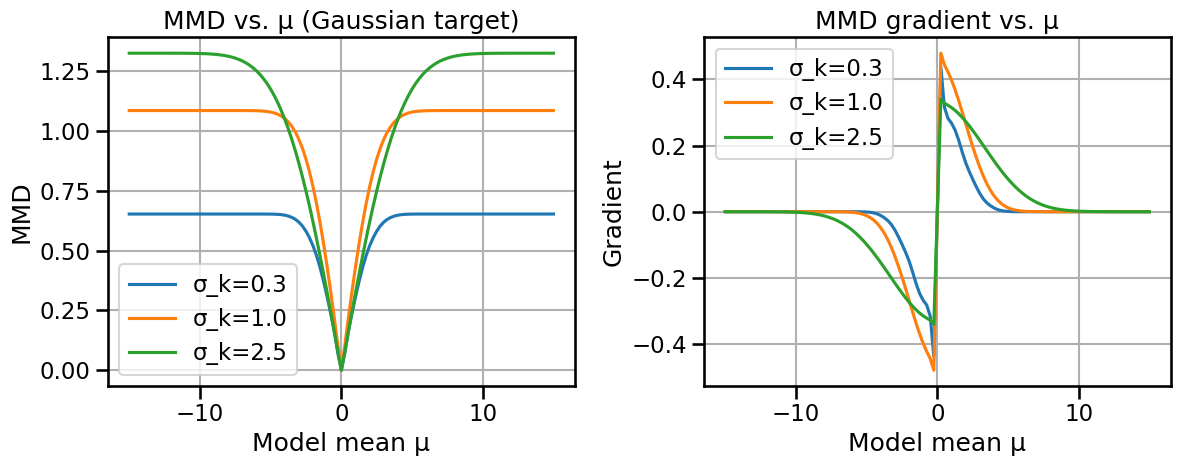
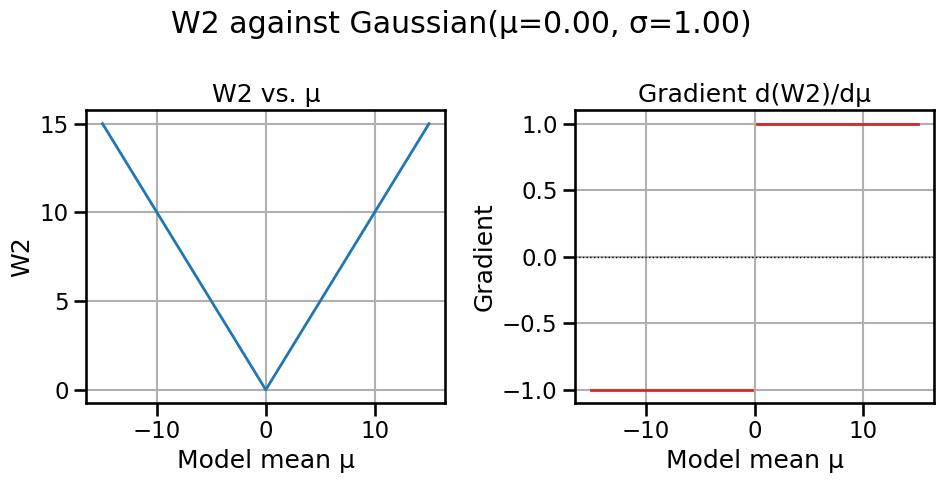
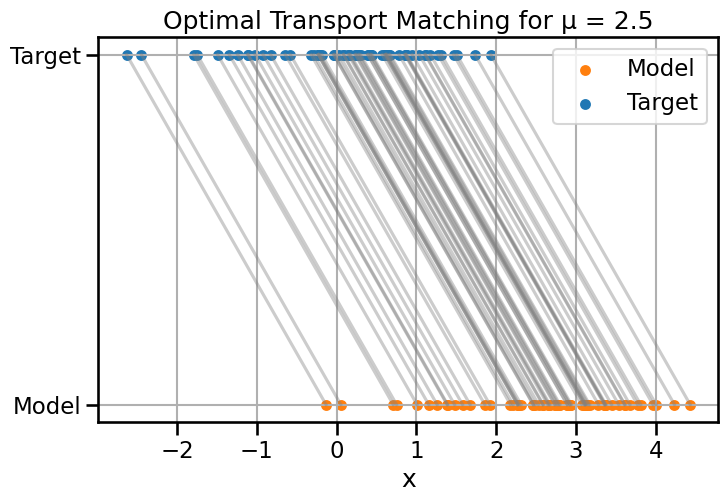
# Compare Wasserstein-2 and Fisher-Rao distances/geodesics
if widgets is None:
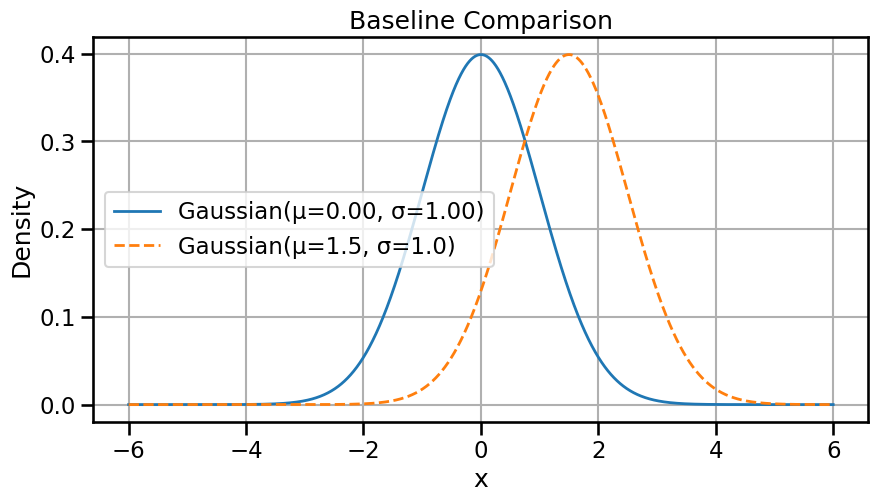
mu1, sigma1 = 0.0, 1.0
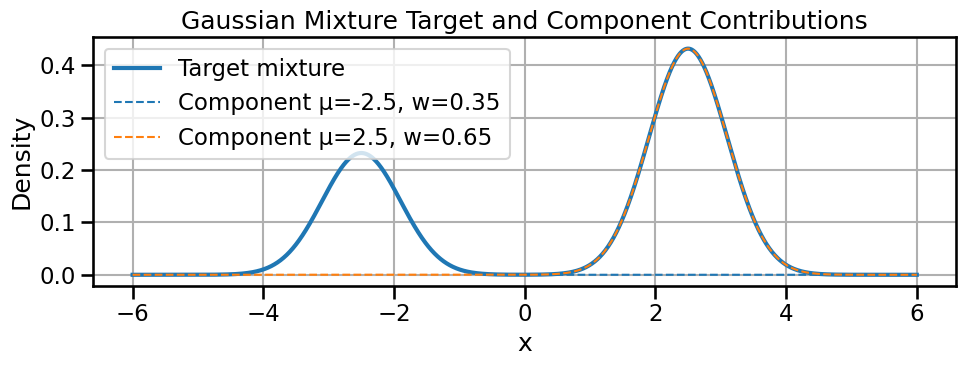
mu2, sigma2 = 2.5, 0.5
w2_val = wasserstein_2(
torch.tensor(mu2, dtype=DEFAULT_DTYPE, device=device),
make_gaussian(mean=mu1, std=sigma1),
model_std=sigma2,
).item()
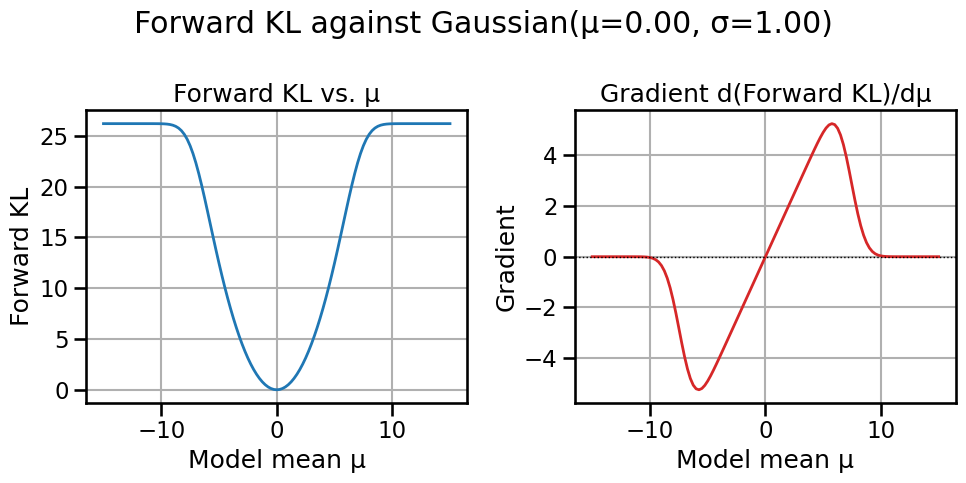
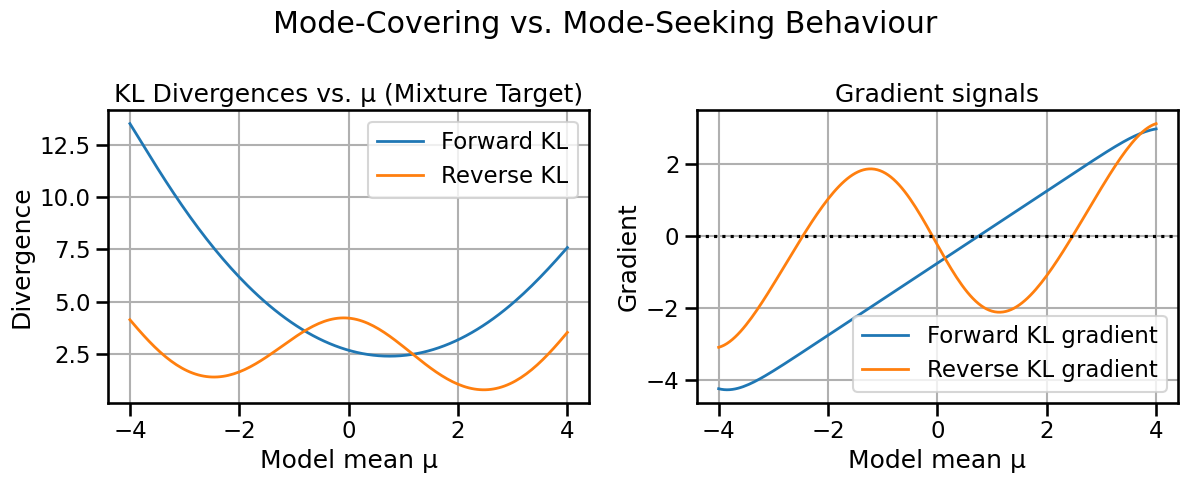
fr_val = fisher_rao_distance(mu1, sigma1, mu2, sigma2)
print(f"W2(p, q) = {w2_val:.3f}\nFisher–Rao(p, q) = {fr_val:.3f}")
ts_ot, mus_ot, sigmas_ot = ot_geodesic(mu1, sigma1, mu2, sigma2, num_points=200)
ts_fr, mus_fr, sigmas_fr = fisher_rao_geodesic(mu1, sigma1, mu2, sigma2, num_points=200)
fig, axes = plt.subplots(1, 2, figsize=(12, 4))
axes[0].plot(ts_ot, mus_ot, label="OT geodesic", linewidth=2)
axes[0].plot(ts_fr, mus_fr, label="Fisher–Rao geodesic", linewidth=2)
axes[0].set_xlabel("t")
axes[0].set_ylabel("Mean μ(t)")
axes[0].set_title("Mean evolution")
axes[0].legend()
axes[1].plot(ts_ot, sigmas_ot, label="OT geodesic", linewidth=2)
axes[1].plot(ts_fr, sigmas_fr, label="Fisher–Rao geodesic", linewidth=2)
axes[1].set_xlabel("t")
axes[1].set_ylabel("Std σ(t)")
axes[1].set_title("Scale evolution")
axes[1].legend()
fig.tight_layout()
plt.show()
sample_ts = np.linspace(0.0, 1.0, 6)
colors = plt.cm.viridis(sample_ts)
grid_np = GRID_X.cpu().numpy()
fig_interp, interp_axes = plt.subplots(1, 2, figsize=(14, 4), sharey=True)
for t_val, color in zip(sample_ts, colors):
mu_t_ot = np.interp(t_val, ts_ot, mus_ot)
sigma_t_ot = np.interp(t_val, ts_ot, sigmas_ot)
pdf_ot = gaussian_pdf_torch(GRID_X, mu_t_ot, sigma_t_ot).cpu().numpy()
interp_axes[0].plot(grid_np, pdf_ot, color=color, linewidth=2, label=f"t={t_val:.2f}")
interp_axes[0].set_title("OT interpolation (W2 geodesic)")
interp_axes[0].set_xlabel("x")
interp_axes[0].set_ylabel("Density")
interp_axes[0].legend(loc="upper right", ncol=2)
for t_val, color in zip(sample_ts, colors):
mu_t_fr = np.interp(t_val, ts_fr, mus_fr)
sigma_t_fr = np.interp(t_val, ts_fr, sigmas_fr)
pdf_fr = gaussian_pdf_torch(GRID_X, mu_t_fr, sigma_t_fr).cpu().numpy()
interp_axes[1].plot(grid_np, pdf_fr, color=color, linewidth=2, label=f"t={t_val:.2f}")
interp_axes[1].set_title("Information-geometry interpolation (Fisher–Rao)")
interp_axes[1].set_xlabel("x")
interp_axes[1].legend(loc="upper right", ncol=2)
fig_interp.tight_layout()
plt.show()
else:
ig_mu1_slider = widgets.FloatSlider(value=0.0, min=-4.0, max=4.0, step=0.1, description="μ₁")
ig_sigma1_slider = widgets.FloatSlider(value=1.0, min=0.2, max=3.0, step=0.05, description="σ₁")
ig_mu2_slider = widgets.FloatSlider(value=2.5, min=-4.0, max=4.0, step=0.1, description="μ₂")
ig_sigma2_slider = widgets.FloatSlider(value=0.5, min=0.2, max=3.0, step=0.05, description="σ₂")
ig_output = widgets.Output()
def _update_geodesic(*_):
with ig_output:
ig_output.clear_output(wait=True)
mu1 = float(ig_mu1_slider.value)
sigma1 = float(ig_sigma1_slider.value)
mu2 = float(ig_mu2_slider.value)
sigma2 = float(ig_sigma2_slider.value)
w2_val = wasserstein_2(
torch.tensor(mu2, dtype=DEFAULT_DTYPE, device=device),
make_gaussian(mean=mu1, std=sigma1),
model_std=sigma2,
).item()
fr_val = fisher_rao_distance(mu1, sigma1, mu2, sigma2)
print(f"W2(p, q) = {w2_val:.3f}\nFisher–Rao(p, q) = {fr_val:.3f}")
ts_ot, mus_ot, sigmas_ot = ot_geodesic(mu1, sigma1, mu2, sigma2, num_points=200)
ts_fr, mus_fr, sigmas_fr = fisher_rao_geodesic(mu1, sigma1, mu2, sigma2, num_points=200)
fig, axes = plt.subplots(1, 2, figsize=(12, 4))
axes[0].plot(ts_ot, mus_ot, label="OT geodesic", linewidth=2)
axes[0].plot(ts_fr, mus_fr, label="Fisher–Rao geodesic", linewidth=2)
axes[0].set_xlabel("t")
axes[0].set_ylabel("Mean μ(t)")
axes[0].set_title("Mean evolution")
axes[0].legend()
axes[1].plot(ts_ot, sigmas_ot, label="OT geodesic", linewidth=2)
axes[1].plot(ts_fr, sigmas_fr, label="Fisher–Rao geodesic", linewidth=2)
axes[1].set_xlabel("t")
axes[1].set_ylabel("Std σ(t)")
axes[1].set_title("Scale evolution")
axes[1].legend()
fig.tight_layout()
plt.show()
sample_ts = np.linspace(0.0, 1.0, 6)
colors = plt.cm.viridis(sample_ts)
grid_np = GRID_X.cpu().numpy()
fig_interp, interp_axes = plt.subplots(1, 2, figsize=(14, 4), sharey=True)
for t_val, color in zip(sample_ts, colors):
mu_t_ot = np.interp(t_val, ts_ot, mus_ot)
sigma_t_ot = np.interp(t_val, ts_ot, sigmas_ot)
pdf_ot = gaussian_pdf_torch(GRID_X, mu_t_ot, sigma_t_ot).cpu().numpy()
interp_axes[0].plot(grid_np, pdf_ot, color=color, linewidth=2, label=f"t={t_val:.2f}")
interp_axes[0].set_title("OT interpolation (W2 geodesic)")
interp_axes[0].set_xlabel("x")
interp_axes[0].set_ylabel("Density")
interp_axes[0].legend(loc="upper right", ncol=2)
for t_val, color in zip(sample_ts, colors):
mu_t_fr = np.interp(t_val, ts_fr, mus_fr)
sigma_t_fr = np.interp(t_val, ts_fr, sigmas_fr)
pdf_fr = gaussian_pdf_torch(GRID_X, mu_t_fr, sigma_t_fr).cpu().numpy()
interp_axes[1].plot(grid_np, pdf_fr, color=color, linewidth=2, label=f"t={t_val:.2f}")
interp_axes[1].set_title("Information-geometry interpolation (Fisher–Rao)")
interp_axes[1].set_xlabel("x")
interp_axes[1].legend(loc="upper right", ncol=2)
fig_interp.tight_layout()
plt.show()
for control in (ig_mu1_slider, ig_sigma1_slider, ig_mu2_slider, ig_sigma2_slider):
control.observe(_update_geodesic, names="value")
_update_geodesic()
controls = widgets.HBox([widgets.VBox([ig_mu1_slider, ig_sigma1_slider]), widgets.VBox([ig_mu2_slider, ig_sigma2_slider])])
maybe_display(widgets.VBox([widgets.HTML("<h4>IG vs. OT Geodesic Explorer</h4>"), controls, ig_output]))