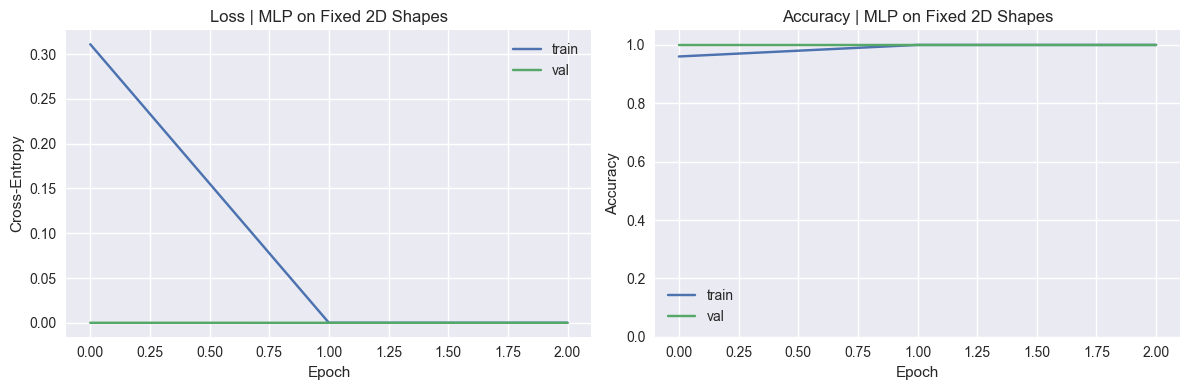
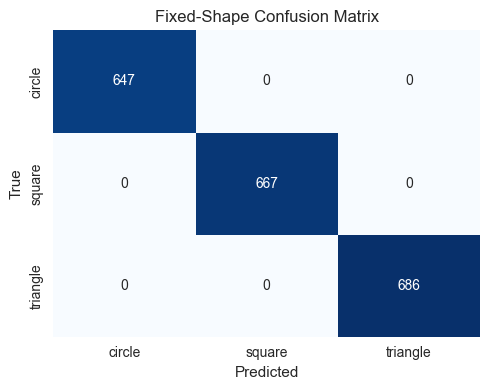
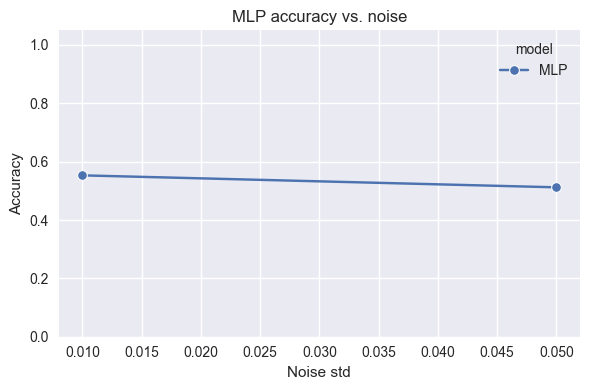
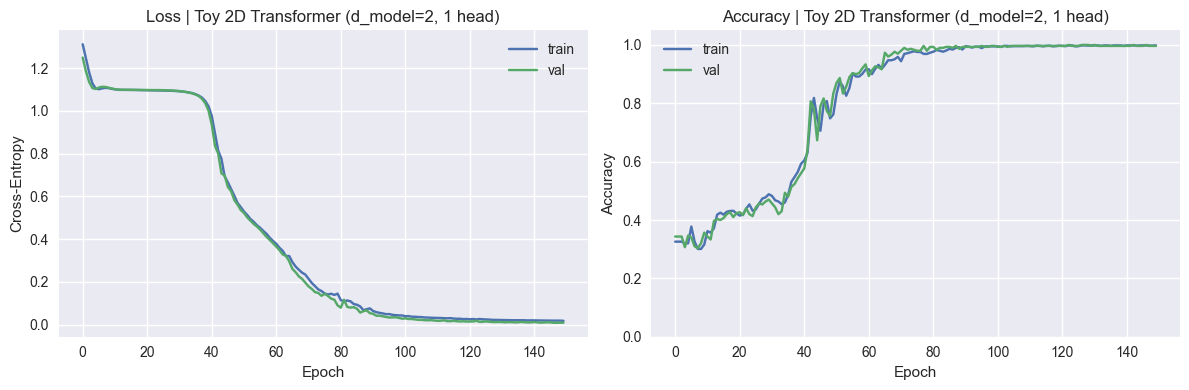
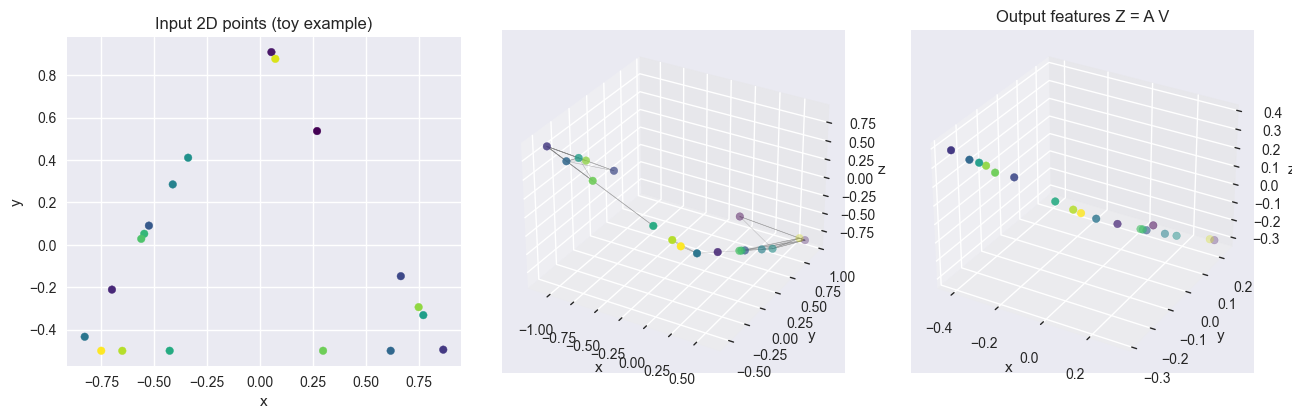
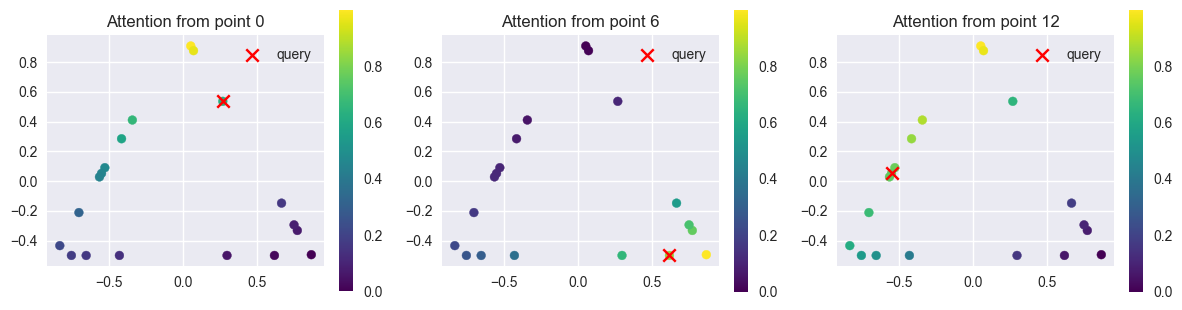
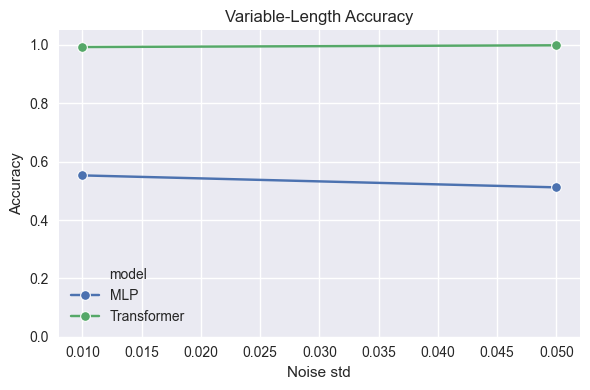
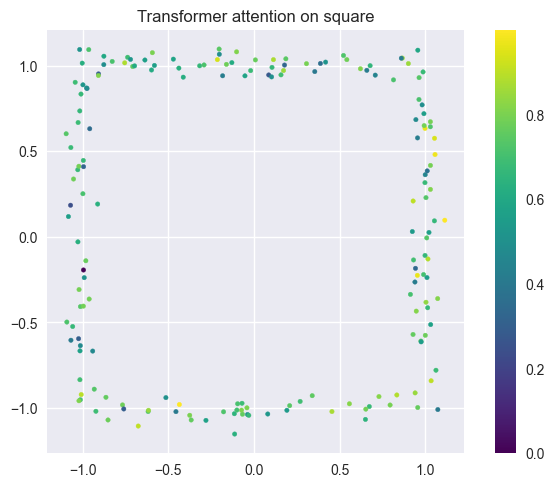
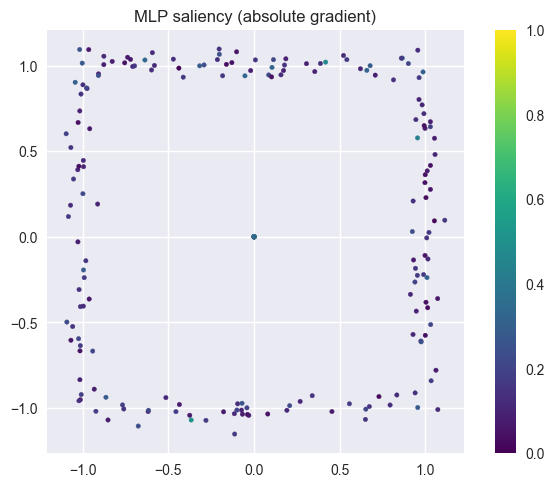
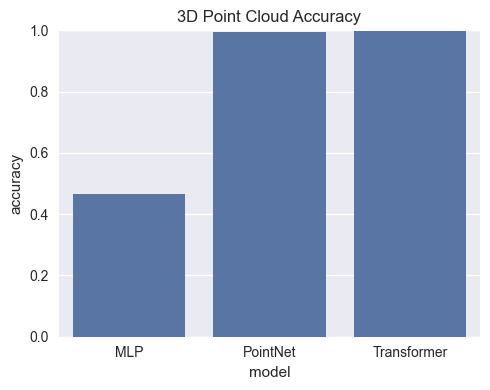
SHAPE_TYPES_3D = ["sphere", "cube", "cylinder", "pyramid"]
def random_rotation_matrix() -> np.ndarray:
angles = np.random.uniform(0, 2 * np.pi, size=3)
cx, cy, cz = np.cos(angles)
sx, sy, sz = np.sin(angles)
rot_x = np.array([[1, 0, 0], [0, cx, -sx], [0, sx, cx]])
rot_y = np.array([[cy, 0, sy], [0, 1, 0], [-sy, 0, cy]])
rot_z = np.array([[cz, -sz, 0], [sz, cz, 0], [0, 0, 1]])
return rot_z @ rot_y @ rot_x
def generate_3d_shape(shape_type: str, n_points: int, noise_std: float = 0.01) -> np.ndarray:
"""Generate 3D shape point cloud with exactly n_points.
Fixes previous cube face broadcasting issue by using scalar axis assignment
with per-face coordinate sampling. Adds safety to enforce exact length.
"""
if shape_type not in SHAPE_TYPES_3D:
raise ValueError(shape_type)
if shape_type == "sphere":
phi = np.random.uniform(0, 2 * np.pi, size=n_points)
costheta = np.random.uniform(-1, 1, size=n_points)
theta = np.arccos(costheta)
x = np.sin(theta) * np.cos(phi)
y = np.sin(theta) * np.sin(phi)
z = np.cos(theta)
points = np.stack([x, y, z], axis=1)
elif shape_type == "cube":
# Choose a face for each point: 0:+x,1:-x,2:+y,3:-y,4:+z,5:-z
faces = np.random.choice(6, size=n_points)
# For each point we need two free coordinates in [-1,1]
free = np.random.uniform(-1, 1, size=(n_points, 2))
points = np.zeros((n_points, 3))
for face in range(6):
mask = faces == face
if not mask.any():
continue
m = mask.sum()
if face == 0: # +x face
points[mask, 0] = 1.0
points[mask, 1] = free[mask, 0]
points[mask, 2] = free[mask, 1]
elif face == 1: # -x face
points[mask, 0] = -1.0
points[mask, 1] = free[mask, 0]
points[mask, 2] = free[mask, 1]
elif face == 2: # +y face
points[mask, 1] = 1.0
points[mask, 0] = free[mask, 0]
points[mask, 2] = free[mask, 1]
elif face == 3: # -y face
points[mask, 1] = -1.0
points[mask, 0] = free[mask, 0]
points[mask, 2] = free[mask, 1]
elif face == 4: # +z face
points[mask, 2] = 1.0
points[mask, 0] = free[mask, 0]
points[mask, 1] = free[mask, 1]
elif face == 5: # -z face
points[mask, 2] = -1.0
points[mask, 0] = free[mask, 0]
points[mask, 1] = free[mask, 1]
elif shape_type == "cylinder":
angles = np.random.uniform(0, 2 * np.pi, size=n_points)
heights = np.random.uniform(-1, 1, size=n_points)
x = np.cos(angles)
y = np.sin(angles)
z = heights
points = np.stack([x, y, z], axis=1)
else: # hollow pyramid with points sampled on each side
# Define pyramid vertices: apex at (0, 0, 1), square base at z = -1
apex = np.array([0.0, 0.0, 1.0])
base_vertices = np.array([
[1.0, 1.0, -1.0],
[-1.0, 1.0, -1.0],
[-1.0, -1.0, -1.0],
[1.0, -1.0, -1.0]
])
# Distribute points across 4 triangular side faces + square base
n_faces = 5
base_count = n_points // n_faces
rem = n_points % n_faces
counts = [base_count + (i < rem) for i in range(n_faces)]
face_points = []
# Sample points on each triangular side face (apex to two base corners)
for i in range(4):
n_face = counts[i]
if n_face == 0:
continue
v1 = base_vertices[i]
v2 = base_vertices[(i + 1) % 4]
# Barycentric coordinates for triangle (apex, v1, v2)
u = np.random.uniform(0, 1, size=n_face)
v = np.random.uniform(0, 1, size=n_face)
# Fold triangle: if u + v > 1, reflect to other half
mask = (u + v) > 1
u[mask] = 1 - u[mask]
v[mask] = 1 - v[mask]
w = 1 - u - v
pts = (w[:, None] * apex +
u[:, None] * v1 +
v[:, None] * v2)
face_points.append(pts)
# Sample points uniformly on the square base face
n_base = counts[4]
if n_base > 0:
# Uniform sampling on the square at z = -1, x in [-1, 1], y in [-1, 1]
base_x = np.random.uniform(-1, 1, size=n_base)
base_y = np.random.uniform(-1, 1, size=n_base)
base_z = np.full(n_base, -1.0)
base_pts = np.stack([base_x, base_y, base_z], axis=1)
face_points.append(base_pts)
points = np.concatenate(face_points, axis=0) if face_points else np.zeros((0, 3))
# Add noise and random rotation
points += np.random.normal(scale=noise_std, size=points.shape)
points = points @ random_rotation_matrix().T
# Safety: enforce exact length (already ensured but keep consistency with 2D generator)
if points.shape[0] != n_points:
if points.shape[0] > n_points:
points = points[:n_points]
else:
extra_idx = np.random.choice(points.shape[0], n_points - points.shape[0], replace=True)
points = np.concatenate([points, points[extra_idx]], axis=0)
return points
class SyntheticPointCloudDataset(Dataset):
def __init__(self, num_samples: int = 3000, length_range: Tuple[int, int] = (200, 1000), noise_std: float = 0.02):
self.samples: List[torch.Tensor] = []
self.labels: List[int] = []
self.max_points = length_range[1]
for _ in range(num_samples):
label = random.randrange(len(SHAPE_TYPES_3D))
n_points = random.randint(*length_range)
pts = generate_3d_shape(SHAPE_TYPES_3D[label], n_points=n_points, noise_std=noise_std)
self.samples.append(torch.tensor(pts, dtype=torch.float32))
self.labels.append(label)
def __len__(self) -> int:
return len(self.labels)
def __getitem__(self, idx: int) -> Dict[str, torch.Tensor]:
return {"points": self.samples[idx], "labels": torch.tensor(self.labels[idx], dtype=torch.long)}
def build_pointcloud_collate(max_points: int, dims: int = 3):
def collate(batch: List[Dict[str, torch.Tensor]]) -> Dict[str, torch.Tensor]:
batch_size = len(batch)
padded = torch.zeros(batch_size, max_points, dims)
mask = torch.ones(batch_size, max_points, dtype=torch.bool)
labels = torch.zeros(batch_size, dtype=torch.long)
for i, item in enumerate(batch):
pts = item["points"]
n = min(pts.size(0), max_points)
padded[i, :n] = pts[:n]
mask[i, :n] = False
labels[i] = item["labels"]
flat = padded.reshape(batch_size, -1)
return {"points": padded, "mask": mask, "inputs": flat, "labels": labels}
return collate